Viral Immunology
Our laboratory is situated in the Henry Wellcome Building at the MRC-University of Glasgow Centre for Virus Research (CVR). Our main interests are the ways in which viruses cause disease; the immune response to infection; and the development of viral vaccines.
Our work focuses on viruses of importance to human and veterinary medicine, including coronaviruses, morbilliviruses, caliciviruses, bunyaviruses, lentiviruses, rhabdoviruses and parvoviruses. Active research projects are investigating:
- development of serological tests for emerging (and re-emerging) viruses
- cross-neutralisation of SARS-CoV-2 variants
- determinants of SARS-CoV-2 virulence, evasion of immunity and vaccine effectiveness
- correlates of immunity to circulating measles virus genotypes
- cross-species transmission and antibody-mediated neutralisation of morbilliviruses
- SARS-CoV-2 spread in the UK domestic cat population
- drivers of bunyavirus emergence in pastoral and agropastoral communities in Africa
Studies in the Viral Immunology group are led by Prof. Brian Willett and Prof. Margaret Hosie
Emerging (and-re-emerging) viruses
With increasing globalisation, UK citizens travel to countries across the globe where tropical diseases are endemic, and upon their return, a proportion of returning travellers may develop rashes, fevers or encephalitides of unknown origin. Diagnosing the causes of such illnesses can be difficult, as only a small number of cases present each year, hence it is not economically viable for hospital diagnostic laboratories to maintain a full panel of diagnostic tests for the diversity of tropical pathogens that exist. In such "returning traveller" cases, diagnostic samples are shipped to the Rare and Imported Pathogens Laboratory (RIPL) at Porton Down, a laboratory that maintains a broad panel of diagnostic tests for the pathogens imported into the UK from tropical countries, for example dengue, Zika or chikungunya viruses. If point of care tests (POCs), or rapid off-the-shelf diagnostic panels were available locally, this would improve disease management and therapy, obviating the need for retrospective public health interventions following a delayed diagnosis.
We are developing a panel of rapid serological tests for viral infections, many of which target pathogens of concern, and which may be increasing with both climate change and globalisation. Using viral pseudotypes we have developed high throughput, neutralisation tests for viruses including the filoviruses Ebola, Marburg and Sudan virus; the togavirus chikungunya virus; the bunyaviruses Rift Valley fever virus, Crimean Congo haemorrhagic fever virus and Puumala, Andes and Hantaan viruses; and a range of coronaviruses from SARS, SARS-CoV-2 and MERS-CoV. These assay systems are suitable for use at BSL-2 and do not require the handling of live viruses. The tests are rapid and, in many cases, the technologies can be transferred to resource limited countries. As we extend this assay panel to cover the suspected causes of rashes, fevers and encephalitides of unknown aetiologies, our aim is to develop off-the-shelf diagnostic tests that will offer clinicians a rapid insight into the exposure history of returning travellers, informing disease management and possible therapeutic interventions. These diagnostic tests also provide a valuable research tool for enhanced, in country surveillance and assessments of vaccine effectiveness. Ongoing studies are examining pathogen exposure in urban and rural cohorts in Malawi and Uganda.
Immunity to SARS-CoV-2
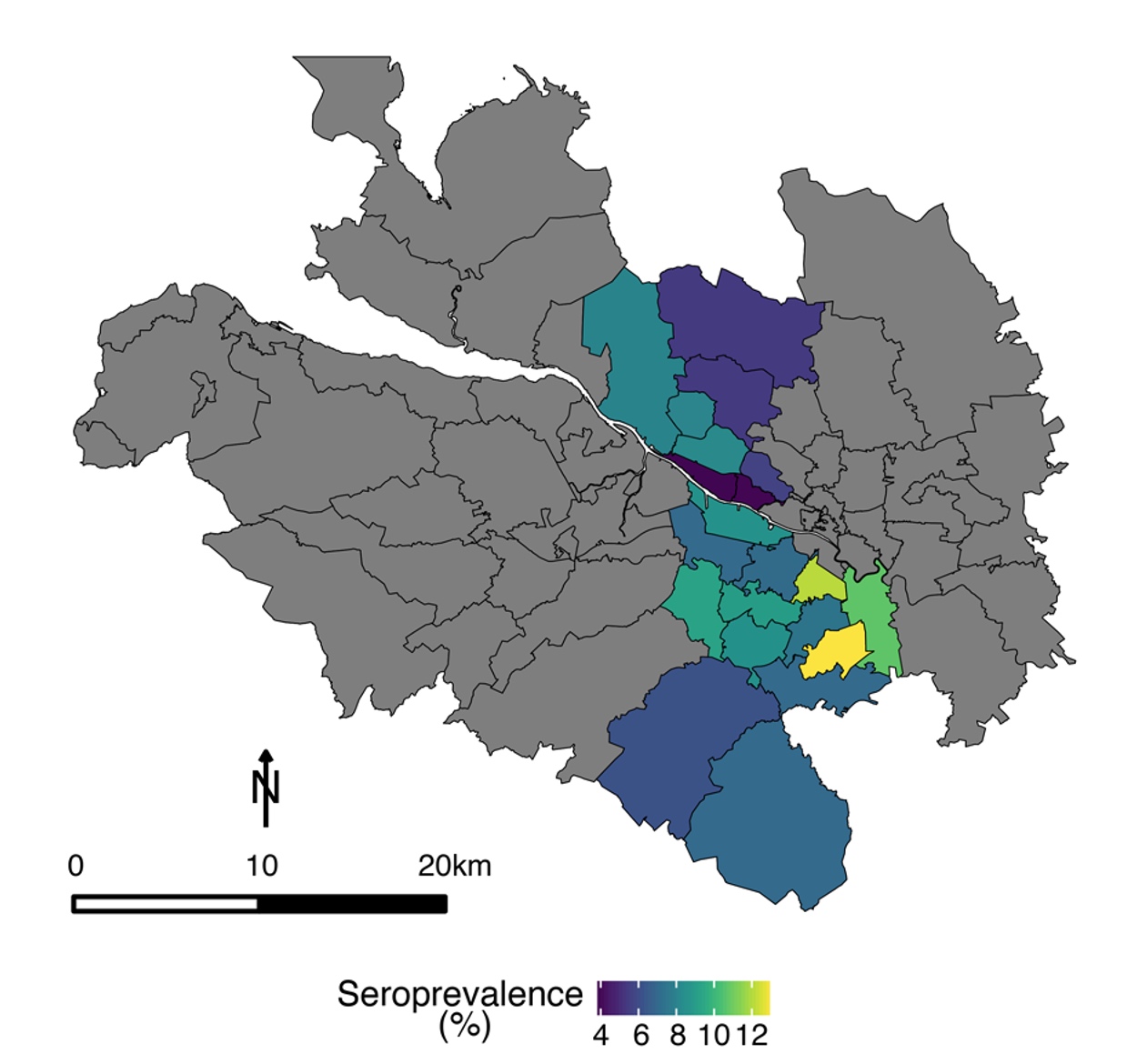
In early 2020, we initiated a program of research aimed at investigating the antibody response to infection with SARS-CoV-2, with a view to tracking the spread of the virus throughout the community and assessing the level of immunity elicited by infection. By combining in-house ELISA assays with pseudotype-based neutralisation assays, we were able to track the spread of SARS-CoV-2 across NHS Greater Glasgow and Clyde, the largest health board in Scotland (Hughes et al. 2021). We showed that males, 45-64-years olds, and patients in secondary care were most likely to be seropositive. Further, we established that approximately half of seropositive individuals (assessed by ELISA) developed neutralising antibodies (Hughes et al. 2021).
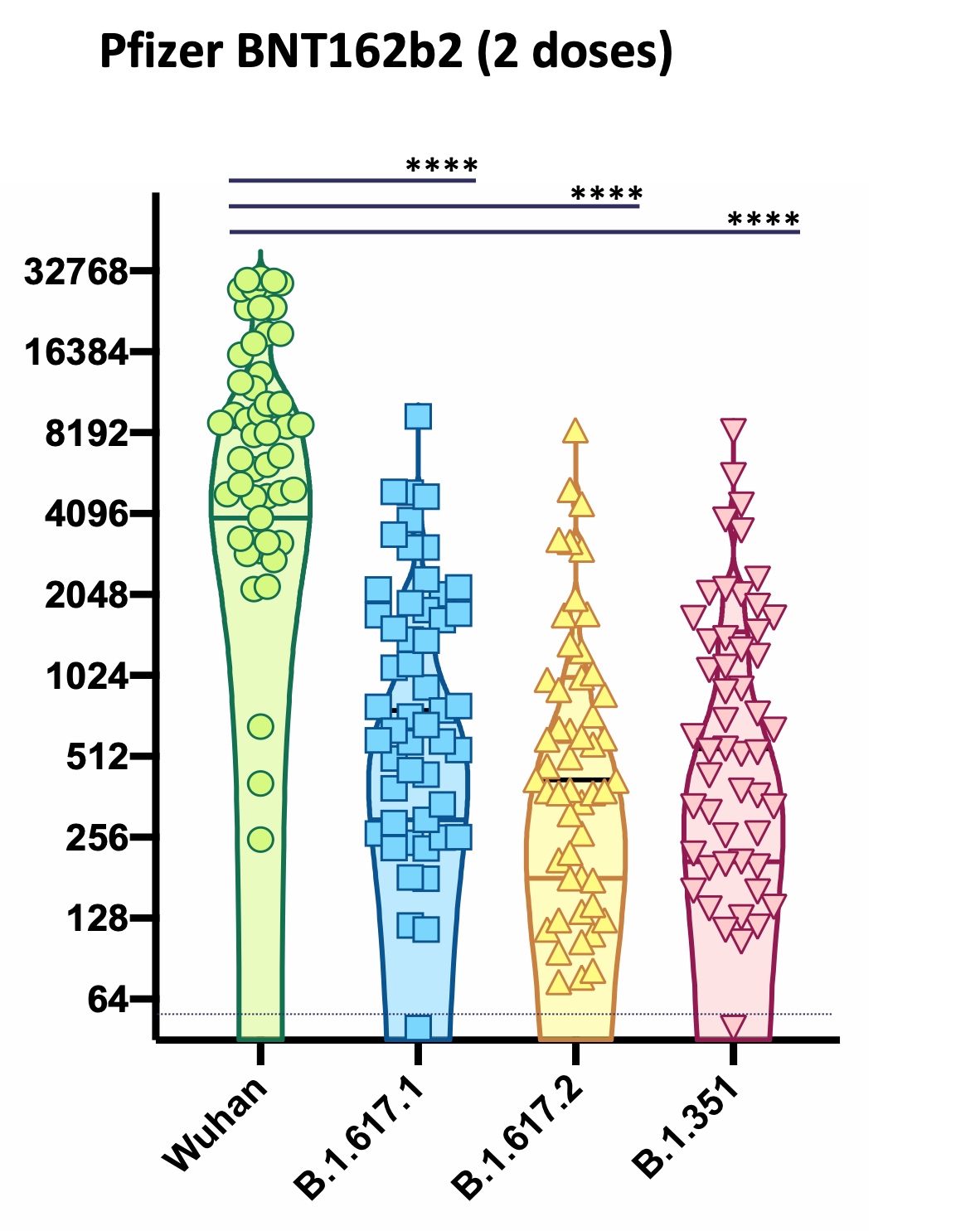 Building on these studies, we examined evasion of the neutralising response by the Beta and Delta variants of SARS-CoV-2 (Davies et al. 2021), and the effect of antigenic variation on evasion of the immune response elicited by vaccination with either ChAdOx1 (AstraZeneca) or BNT162b2 (Pfizer) vaccines. The emergence of the Omicron lineage of viruses at the end of 2021 signalled a new phase in the evolution of SARS-CoV-2.
Building on these studies, we examined evasion of the neutralising response by the Beta and Delta variants of SARS-CoV-2 (Davies et al. 2021), and the effect of antigenic variation on evasion of the immune response elicited by vaccination with either ChAdOx1 (AstraZeneca) or BNT162b2 (Pfizer) vaccines. The emergence of the Omicron lineage of viruses at the end of 2021 signalled a new phase in the evolution of SARS-CoV-2.
Working closely with colleagues across the University of Glasgow and the wider UK academic and healthcare communities, we established that the Omicron variant had a phenotype that had changed fundamentally compared with all prior variants (Research briefing). The Omicron variants displayed reduced neutralisation by antibodies elicited by vaccination, while T cell responses were relatively preserved (Willett et al. 2022). Further, real-world vaccine effectiveness was partially restored by booster vaccination. Our study revealed that Omicron displayed distinct entry and fusion properties to preceding variants, entering cells via a TMPRSS2-independent, endosomal entry pathway (Willett et al. 2022). This fundamental change in the viral entry mechanism was not predicted by sequencing data; rather it was revealed by observations in cell culture experiments from several laboratories, including those at the CVR. This project was supported in part by HDR-UK, further details are available here.
One of our SARS-CoV-2 research objectives is to elucidate the nature of the virus-host interaction; investigating the strength, specificity and duration of humoral immunity. Working closely with colleagues at the University of Birmingham, we are currently investigating vaccine effectiveness in at-risk populations; including children (Dowell et al, 2022a & Dowell et al., 2022b), the elderly (Parry et al, 2022; Parry et al, 2023) and patients with chronic lymphocytic leukemia (Parry et al, 2022a & Parry et al, 2022b). In at-risk populations, the immune response to infection and vaccination may be weaker. As a result, people in this category may require multiple vaccinations to achieve the same level of immunity as the general population.
As the COVID-19 pandemic has ended, the global focus has shifted from "pandemic response" to "long-term management". As nationwide serosurveillance studies have ended, our understanding of the level of immunity in the general population has diminished. While vaccination campaigns have sought to maintain immunity in at-risk populations, the majority of the population are no longer being offered COVID-19 vaccination. Hence immunity is now being elicited through exposure to successive waves of infection with SARS-CoV-2 variants as they circulate as endemic, seasonal coronaviruses. Accordingly, most people now have "hybrid immunity", generated through a combination of vaccinations and infections. Understanding the complex immunological landscape of the general population will be crucial to formulating COVID-19 vaccination strategies going forward. In Viral Immunology, we are measuring the level of immunity in the general population, comparing this with the immunity observed in multiply-vaccinated, at-risk individuals, tracking the level of immune evasion by emerging variants and assessing the effectiveness of novel vaccine formulations as they are introduced.
Emerging morbilliviruses of wildlife and humans
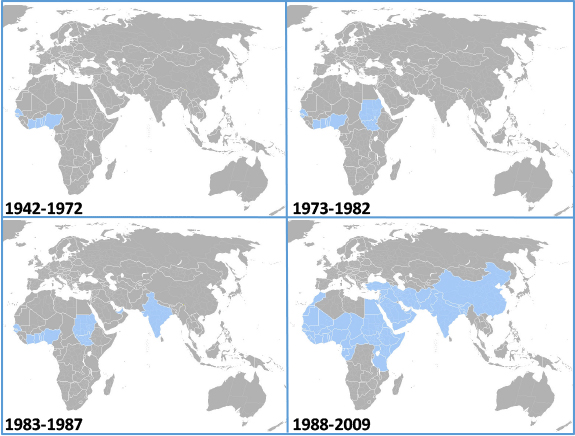
The global spread of peste des petits ruminants virus (PPRV)
In May 2011, the General Session of the Office International des Epizooties declared the world to be free from rinderpest virus, the causative agent of “cattle plague”. Rinderpest virus (RPV) is a morbillivirus, a close relative of measles virus (MeV). Like rinderpest virus, measles virus is now being considered for global eradication by vaccination. However, there is increasing concern that if such a vaccination programme was to be successful, the requirement for vaccination would cease. As a result, humans would no longer have cross-protection against zoonotic infections with closely-related animal morbilliviruses. Significant concerns have now been raised about the threat from the carnivore morbillivirus, canine distemper virus (CDV), a virus capable of infecting diverse species including dogs, ferrets, martens, lions, hyenas and seals. This ability to cross species renders CDV a significant threat to many endangered species of wildlife. Moreover, pathogenic CDV infections have been described in primates, raising the possibility of zoonotic transmissions to humans. There is now intense interest in the viral reservoirs of CDV and the degree of cross-protection conferred by MV vaccination as a means of guarding against cross-species transmission.
Neutralisation of morbilliviruses by antibodies is currently measured using a restricted subset of live viruses that grow readily in culture. Thus it is not possible to compare antibody responses to vaccine viruses with those against primary field strains of virus, making predictions of vaccine efficacy and likelihood of zoonotic transmission challenging. We have developed novel viral pseudotype-based assay systems with which serological responses to diverse animal morbilliviruses can be measured rapidly, with high sensitivity and specificity (see Logan et al. 2016a & Logan et al., 2016b). Using this simple methodology, we are measuring neutralizing antibody responses against primary field strains of virus, novel emerging morbilliviruses, and unique biotypes and serotypes of virus for which assays are currently unavailable. We have developed parallel systems for diverse morbilliviruses; viral pseudotypes based on peste des petits ruminants (PPRV), CDV, RPV, MeV and phocine distemper virus (PDV), and target cell lines expressing the cognate receptors for each virus.
This project brings a comprehensive understanding of the spread of morbilliviruses in livestock (Herzog et al., 2019; 2020a & 2020b) and wildlife species (Gilbert et al., 2020). It addresses whether morbilliviruses are being maintained in atypical host species and whether these species are a source of infection for susceptible hosts. It provides epidemiological and biological data to inform future strategies for virus eradication by vaccination, illuminating the extent of virus spread and the likely species that will require to be vaccinated. The high-throughput assay techniques we employ build capacity in the UK for "rapid response" to emerging viral diseases.
The importance of emerging morbilliviruses to animal health is exemplified by the recent outbreaks of PPRV in Greece, Romania and Bulgaria. In July 2024, peste des petits ruminants (PPR) was detected for the first time in Greece and Romania, both previously officially free of the disease. As of 9 August 2024, Greece had reported 47 outbreaks, with over 2000 cases, while Romania notified 56 outbreaks, with over 5000 cases, through the World Animal Health Information System (WAHIS). Ongoing epidemiological and laboratory investigations aim to understand the transmission pathways and extent of virus circulation in the affected countries. Such outbreaks have a significant economic impact of the affected countries, and the livelihoods of farmers, with immediate export bans on dairy products, culling in affected flocks and herds, and vaccination of animals in surrounding areas. At the CVR, the Viral Immunology laboratory has developed novel approaches to serosurveillance, enabling researchers to assess the level of spreas of PPRV and related morbilliviruses in both livestock and wildlife.
Listen to Prof. Willett discussing morbillivirus research at the CVR here: CVR Podcast
Research highlights: the emergence of SARS-CoV-2 Omicron
Omicron: a novel SARS-CoV-2 phenotype and antigenicity
The Omicron variant of SARS-CoV-2 had a biological phenotype that had changed fundamentally from the preceding variants derived from the ancestral "Wuhan-Hu-1" virus (lineage B.1). Multiple mutations in the spike protein of the virus resulted in a virus with a novel antigenicity, a virus that was neutralised poorly by sera elicited against infection with preceding lineages, or against vaccines based upon the protypic Wuhan-Hu-1 strain of virus. The Omicron variant acquired a new endocytic mechanism of infection, altering the cell tropism of the virus and decreasing viral virulence.
Background
When the Omicron variant of SARS-CoV-2 was first identified in South Africa, there was significant apprehension globally about the potential threat of a new wave of infection. Fears stemmed from the abundance of mutations that had been acquired during the evolution of Omicron, would they affect the ability of vaccines to prevent symptoms of disease? How immune evasive would the novel viral variant be? While the Omicron variant spread rapidly from country to country, outpacing the preceding Delta variant, data were required urgently to define the biological properties of the virus and the likely effects of the antigenic change in the spike protein on real-world vaccine effectiveness.
The DOVE study
Neutralising antibodies and cytotoxic T cells targeting the spike protein of the Omicron variant were quantified in samples from recipients of deployed vaccines (ChAdOx1, BNT162b2 and mRNA-1273) in the COVID-19 Deployed Vaccine Cohort Study (DOVE). Vaccine effectiveness was estimated in the Evaluation of Variants Affecting Deployed COVID-19 Vaccine (EVADE) Study that linked vaccine responses with genetic variants in Scotland. As suggested by the ‘early warning’ signal from sequencing data that predicted substantial immune escape, we found markedly reduced neutralisation of Omicron BA.1 and BA.2 variants by sera from recipients of two or three doses of vaccine, while T cell responses were relatively preserved. These data were mirrored by a substantial reduction in real-world vaccine effectiveness, which was partially restored by booster vaccination.
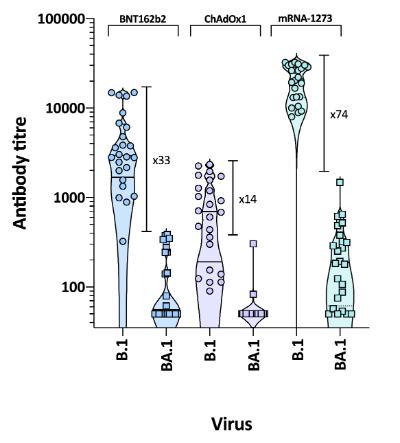
Figure 1. Neutralisation of ancestral (B.1) and Omicron variant (BA.1) viruses by vaccine sera. Sera elicited by vaccination with two doses of BNT162b2 (Pfizer), ChAdOx1 (AstraZeneca) or mRNA-1273 (Moderna) were screened for neutralising activity against viral pseudotypes bearing the spike proteins of either the ancestral (vaccine) virus (B.1) or the Omicron variant (BA.1). Neutralising activity against Omicron was significantly lower than that against B.1. However, as recipients of the Pfizer and Moderna mRNA-based vaccines had higher levels of neutralising activity against B.1, they retained greater cross-neutralising activity against the novel, Omicron variant.
Novel biological properties of the Omicron variant
We studied the virological properties of Omicron in vitro and found that the novel SARS-CoV-2 variant did not induce cell-cell fusion, a phenomenon observed in preceding variants. Cell-cell fusion results from the activation of spike protein at the cell membrane by the cell surface protease TMPRSS2. Unlike preceding variants, Omicron variants entered cells preferentially via a TMPRSS2-independent endosomal entry pathway, thereby altering the cell tropism of the virus.
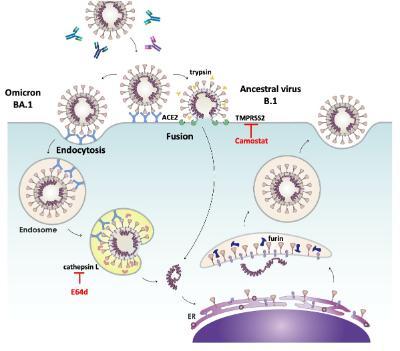
Figure 2. SARS-CoV-2 entry routes. The ancestral virus (B.1) and subsequent variants (Alpha, Delta etc) entered cells predominantly by fusion at the plasma membrane, a process triggered by the protease TMPRSS2 and which could be inhibited by Camostat. In contrast, the Omicron variant displayed an enhanced ability to infect via an endocytic route that was dependent on the protease cathepsin L. This endocytic route could be blocked by the cathepsin inhibitor E64d. Hence, in cell types where TMPRSS2 was not available, Omicron could infect more efficiently than ancestral viruses, by using an endocytic entry route. Image adapted from Tang et al, 2020.
Experiments using chimaeric spike proteins revealed that impaired cell-cell fusion was determined by changes within the receptor binding domain of the spike protein, whilst endosomal entry was associated with changes in the S2 domain of the protein. Such major changes in antigenicity and replicative biology of the virus likely contributed significantly to the global spread and decreased virulence of the Omicron variant.
The findings from our studies on the Omicron variant are published here Willett et al, 2022.
Research highlights: viral receptors
The primary receptor for FIV
The initial event in the process of viral infection involves the binding of the virus to a molecule on the cell surface. Therefore, the expression pattern of this molecule within the host determines which cell types the virus will target. The receptor for human immunodeficiency virus (HIV) is CD4, a molecule that is expressed primarily on a specialised subset of cells within the immune system known as helper T cells (Th cells). Th cells play a pivotal role in the development of specific immune responses to pathogens and by targeting and destroying these cells selectively, HIV impairs the ability of the immune system to respond to infection. Consequently, HIV-infected individuals develop opportunistic infections and ultimately AIDS (acquired immune deficiency syndrome).
Although FIV targets Th cells, it has been known for several years that the virus does not use CD4 as a primary receptor. In a recent study, published in the journal Science, researchers from the Retrovirus Research Laboratory joined with colleagues in London and Japan to show that the primary receptor for FIV was a molecule known as CD134 (OX40).
What is CD134?
CD134 belongs to a large group of related molecules known as the nerve growth factor (NGF)/tumour necrosis factor (TNF) receptor superfamily. The molecule was first identified as "OX40", an antigen expressed on the surface of rat Th cells. It was subsequently found to be identical to an activation antigen found on human T cells known as "BerACT35".
The molecule functions in the development of antigen-specific T cell responses where it supports T cell expansion and survival by interacting with its ligand, CD134L (OX40L) on antigen presenting cells. Expression of feline CD134 expression on non-susceptible cells (eg human cells) renders the cells permissive for infection with FIV. Infection requires co-expression of a second molecule, CXCR4, a molecule we identified previously as the major co-receptor for FIV (see Nature 1997 J. Virology 1997).
Group members

Diagnostic tools
Monoclonal antibodies
Many of the antibodies generated in our laboratory can be purchased from Bio-Rad. The current list of reagents available is as follows:
Feline leucocyte differentiation antigens
|
Specificity |
Designation |
Catalogue number |
Applications |
| CD134 | 7D6 | MCA2568 | FC,WB |
| CD134-FITC | 7D6 | MCA2568F | FC |
| CD134-A488 | 7D6 | MCA2568A488 | FC |
| CD134-A647 | 7D6 | MCA2568A647 | FC |
|
CD4 |
vpg34 |
MCA1346 |
FC, IP |
|
CD4-FITC* |
vpg34 |
MCA1346F |
FC |
|
CD4 |
vpg38 |
MCA1350 |
FC, IP |
|
CD4 |
vpg39 |
MCA1349 |
FC, IP |
|
CD8-RPE* |
vpg9 |
MCA1347PE |
FC |
|
CD9 |
vpg15 |
MCA1345 |
FC, IP |
|
MHC class II |
vpg3 |
MCA1348 |
FC, IP |
*CD4-FITC and CD8-RPE may be used for dual colour analysis of feline T cell subsets in a single tube format. The histograms below represent an analysis of T cell subsets in an FIV-infected cat using vpg34-FITC and vpg9-PE. Note the appearance of the "CD8low" sub-population (Immunology (1993), 78:1-6), a sub-population of CD8+ T cells thought to be CD8a + b low (J Gen Virol. (1998), 79:91-94). This population is often observed in FIV infected cats.
Feline Immunodeficiency Virus
|
Specificity |
Designation | Catalogue number | Applications |
| gp120 | vpg68 | MCA1351 | IFA, FC, IP |
| p24 gag | vpg50 | MCA1353 | IFA, IP, WB |
Glutathione-S-Transferase
| Specificity | Designation |
Catalogue number |
Applications |
| GST | vpg66 | MCA1352 | WB, IP |
| GST-biotinylated | vpg66 | MCA1352B | WB, IP |
vpg66 will recognise fusion proteins generated using the pGEX-2T vector and is ideal for both immunoprecipitation and western blot analyses. The use of vpg66 to detect fusion proteins between the HIV-1 Nef protein and GST is detailed in Harris, M. and Coates, K.C. (1993). J. Gen. Virol. 74, 1581-1589 and Harris, M.P.G. and Neil, J.C. (1994). J. Mol. Biol. 241, 136-142.
Additional antibodies against feline leukaemia virus, distinct epitopes on feline CD4, FIV p24 or FIV gp120 are available. Further information can be obtained from either our laboratory or Bio-Rad.

